This is a joint venture between Philips Research Suresnes FRANCE, CREATIS and INRIA (Team ASCLEPIOS).
Overview
Quantification of cardiac deformation and strain through different image modalities (3D ultrasound and magnetic resonance imaging being the most widely used techniques) has been the object of intense research. However, a widespread use of these techniques in clinical routine is still held back due to the lack of strong studies which accuratly quantify and compare the performance of existing methods in each modality. The use of synthetic sequences is one of the most promising tool for advanced initial in silico evaluation. Nevertheless, the realism of existing simulation techniques still needs to be improved to represent reliable benchmarking data.
Objectives
We propose in this study the first simulation framework which allows the generation of realistic 3D synthetic cardiac ultrasound and magnetic resonance (both cine and tagged) image sequences from the same virtual patient.
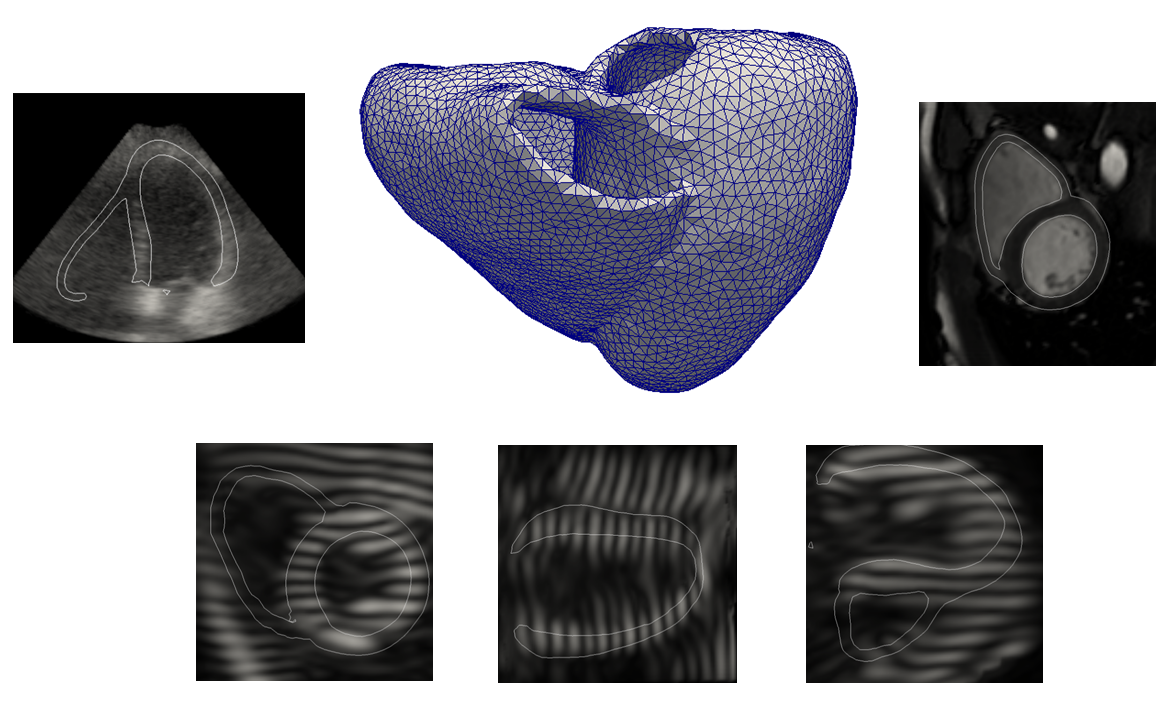
Methodology
A state-of-the-art electromechanical model is combined with physical simulators (one for each modality) in an novel framework which efficiently merges several warping strategies to keep full control of myocardial deformations while preserving realistic image texture. The novelties introduced in this paper are the following :
- The combination of an electromechanical model with a magnetic resonance physical simulator to introduce the possibility of interacting with the image formation process to introduce complex effects;
- The use of multi-modal template sequences from the same patient to extract the most relevant information from each modality in order to generate both cine and tagged synthetic sequences;
- The introduction of a novel combinative warping strategy, with the goal of reducing motion artifacts that may occur in myocardial regions;
- The contribution of the first unified framework which allows the generation of multi-modal (US, cine and tagged MR) realistic synthetic sequences for the same virtual patient.
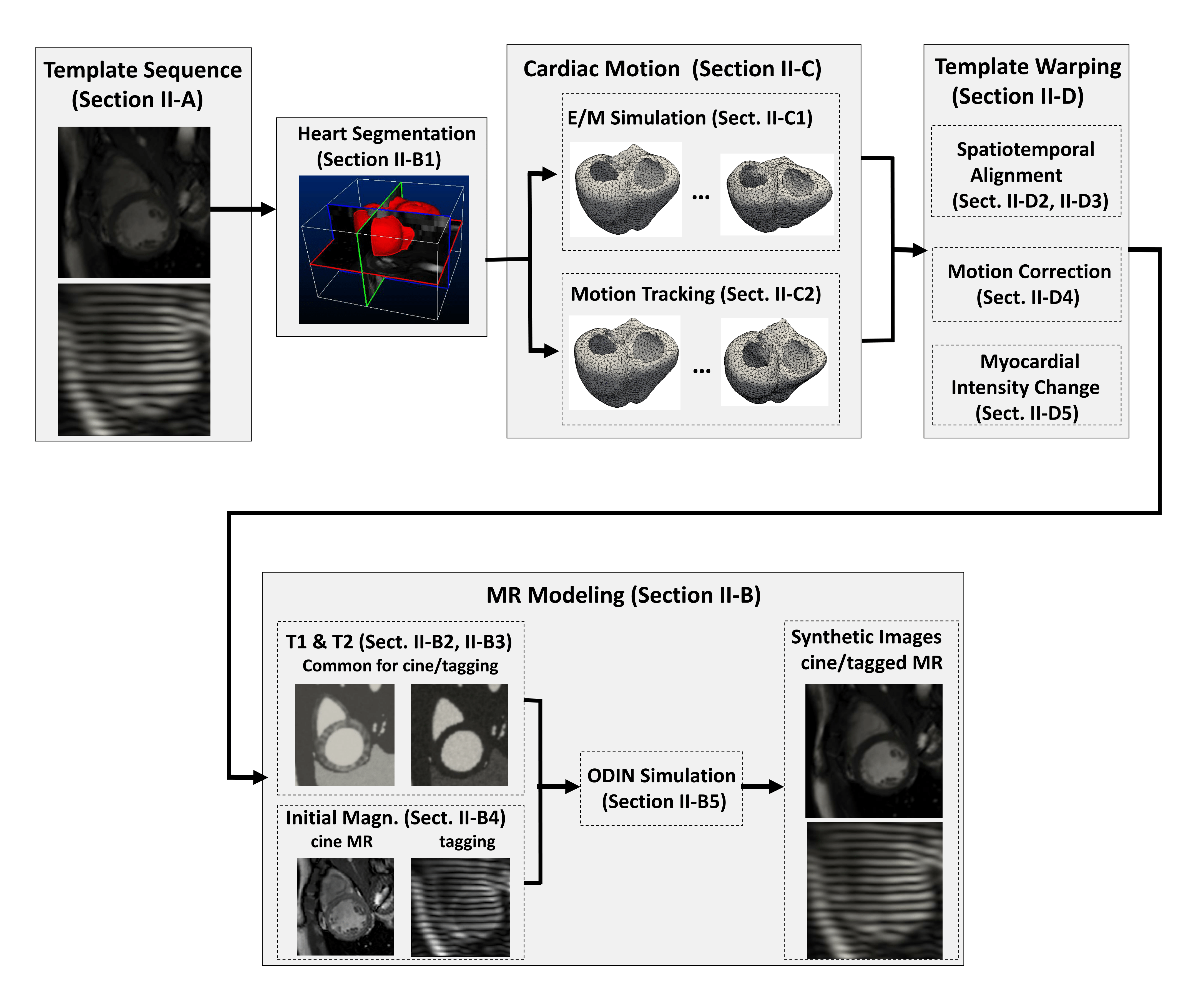
A schematic of the extended pipeline to magnetic resonnance simulation is summarized hereunder
Open-access database
The proposed generic framework was used to generate the first database composed of 3D ultrasound, cine and tagged MR sequences for each of the 18 virtual patients, including healthy and pathological cases. We simulated a total of 90 sequences representing 2700 image volumes.
All the simulations were launched from the virtual imaging platform (VIP) which allows the execution of the simulator as a web service and benefits from the EGI computing power. The generation of one full 3D sequence took around 6 hours on VIP, knowing that would have taken over 300 hours on a personal laptop
Get Started
To browse through the image database, simply connect to the Multimodality STRAUS database, explore and download the images of interest or the entire collection. This database is public, so no login is required.
Download
You can download the entire collection of images or specific images.
Download Collection (Option 1)
Click on the icon menu Collection actions in the upper-right of the collection's header and select Download collection.
Download Collection (Option 2)
Click on the icon Collection actions in the upper-right corner of the table's header and Download collection.
Download Selected Folders
Select the folders of interest and click on the icon Checked actions and Download checked resources.
Each virtual patient folder is organized as follows
| Folder | Content Description |
|---|---|
| cine | 3D cine-MR simulation sequences (raw/mhd) with reference meshes (vtk) |
| tag1 | 3D tag-MR (ch1) simulation sequences (raw/mhd) with reference meshes (vtk) |
| tag2 | 3D tag-MR (ch2) simulation sequences (raw/mhd) with reference meshes (vtk) |
| tag3 | 3D tag-MR (ch3) sequences (raw/mhd) with reference meshes (vtk) |
| us | 3D Ultrasound simulation sequences (raw/mhd) with reference meshes (vtk) |
R&D Team
| Olivier BERNARD | Associate Professor, CREATIS, France
|
| Mathieu De Craene | Research Engineer, Philips Research, Suresnes, France
|
| Maxime Sermesant | Researcher, INRIA, France
|
| Jan D'hooge | Professor, MIRC laboratory, Belgium
|
| Sorina Camarasu-Pop | Research Engineer, CREATIS, France
|
| Martino Alessandrini | Postdoctoral fellow, University of Bologna, Italy
|
| Denis Friboulet | Professor, CREATIS, France
|
| Yitian Zhou | PhD student, Philips Research, Suresnes, France
|
| Sophie Giffard-Roisin | PhD student, INRIA, France
|











