Echocardiography Segmentation with Enforced Temporal Consistency
- This is a joint venture between CREATIS and VITALab laboratories.
- The code is available on github through the following repo.
- The dataset is available at the following warehouse.
- Please refer to our paper for any use of the data or code.
Overview
Convolutional neural networks (CNN) have demonstrated their ability to segment 2D cardiac ultrasound images. However, despite recent successes according to which the intra-observer variability on end-diastole and end-systole images has been reached, CNNs still struggle to leverage temporal information to provide accurate and temporally consistent segmentation maps across the whole cycle. Such consistency is required to accurately describe the cardiac function, a necessary step in diagnosing many cardiovascular diseases.
Objectives
We propose a framework to learn the 2D+time apical long-axis cardiac shape such that the segmented sequences can benefit from temporal and anatomical consistency constraints. Our method is a post-processing that takes as input segmented echocardiographic sequences produced by any state-of-the-art method and processes it in two steps to (i) identify spatio-temporal inconsistencies according to the overall dynamics of the cardiac sequence and (ii) correct the inconsistencies. The identification and correction of cardiac inconsistencies relies on a constrained autoencoder trained to learn a physiologically interpretable embedding of cardiac shapes, where we can both detect and fix anomalies.
Main contributions
The main contributions achieved in this project are:
- We define clinically interpretable indicators to quantitatively evaluate the temporal consistency of 2D+time segmentations;
- We introduce a generic post-processing algorithm, based on an interpretable embedding of cardiac shapes, which can be plugged at the end of any segmentation method, which enforces temporal consistency on top of improving the overall accuracy of the segmentations;
- We make public a new fully-annotated dataset of 98 full cycle apical 4 chamber (A4C) sequences from the CAMUS dataset. Up until now, only ED and ES expert annotations were available for these US sequences. As far as we know, this is the first public dataset of its kind for 2D echocardiograph.
Temporal Echocardiography Dataset
Open access dataset
Some examples of the annotated sequences. Don't hesitate to download each animation by right clicking on it for a better visualization.
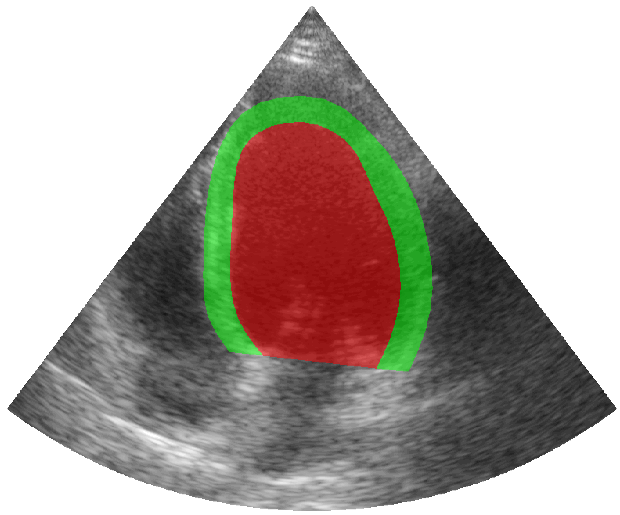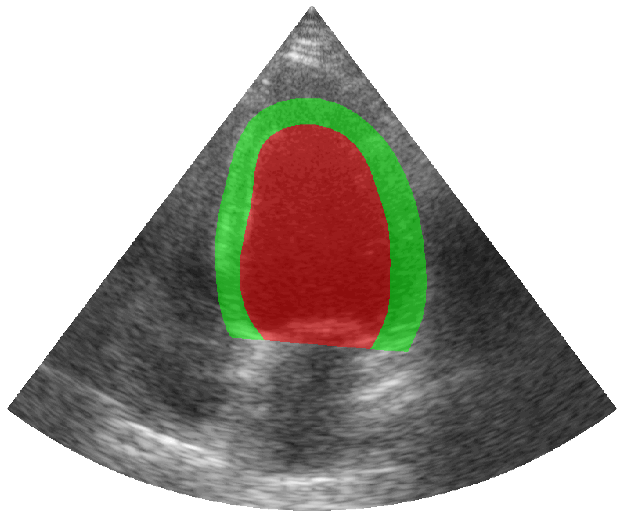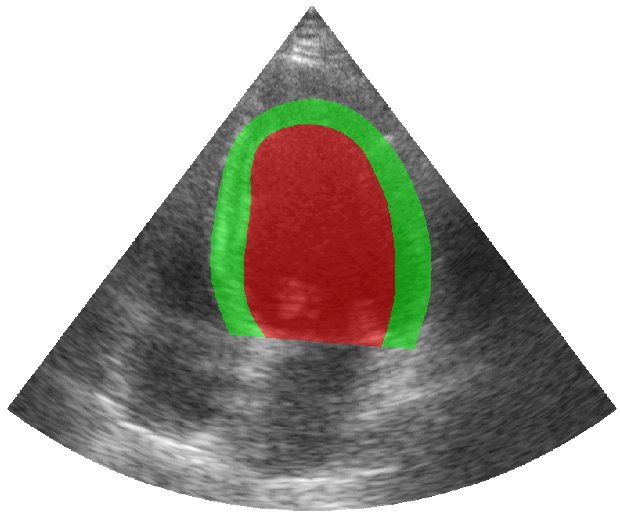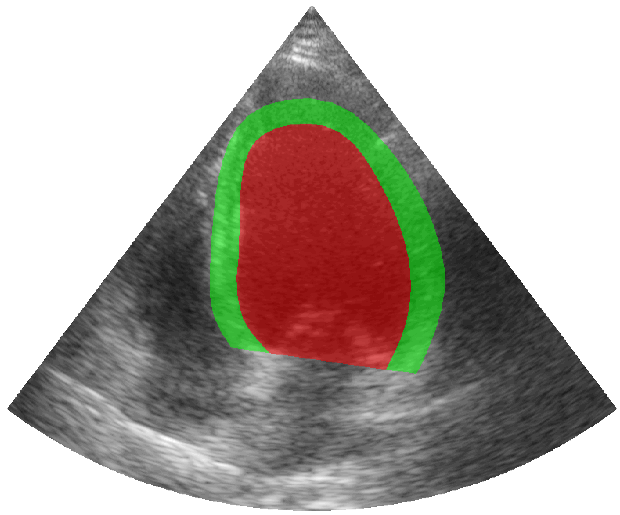
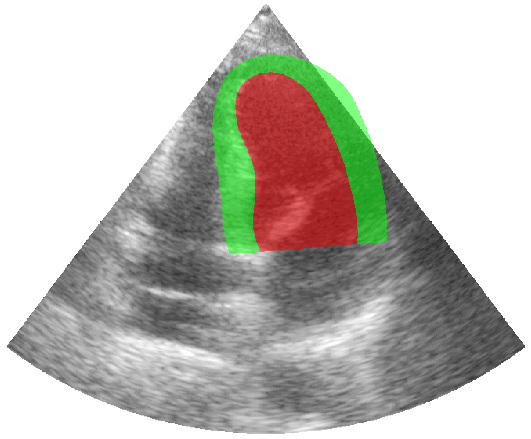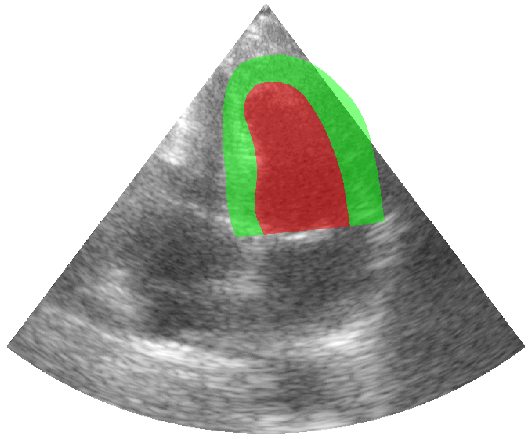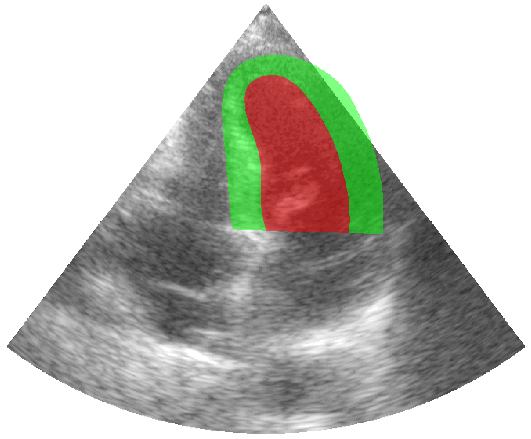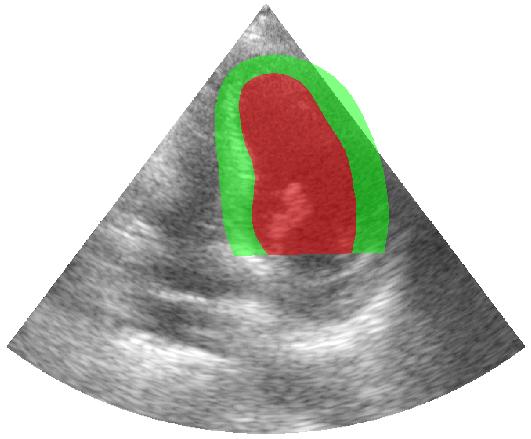
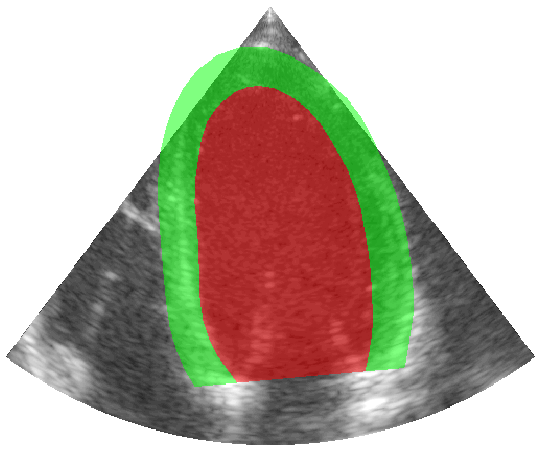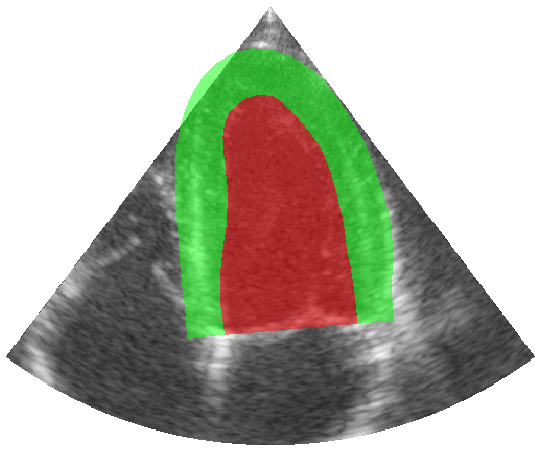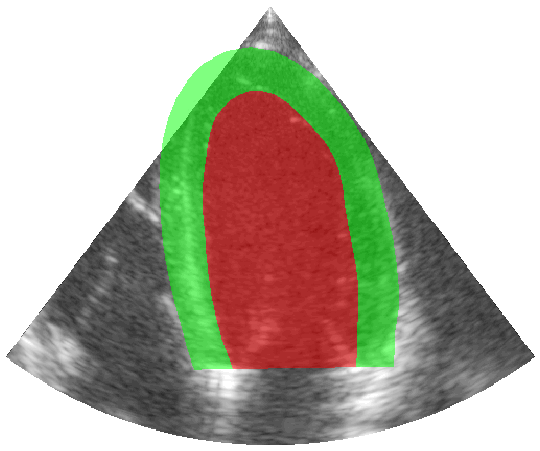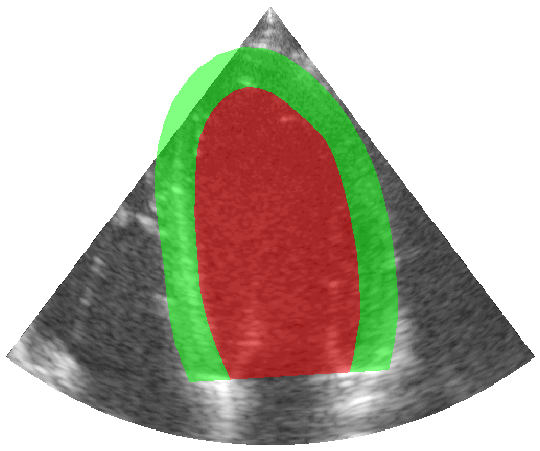
Get Started
To browse through the image database, simply connect to the TED database, explore and download the images of interest or the entire collection. This database is public, so no login is required.
Enforced Temporal Consistency
Overall scheme
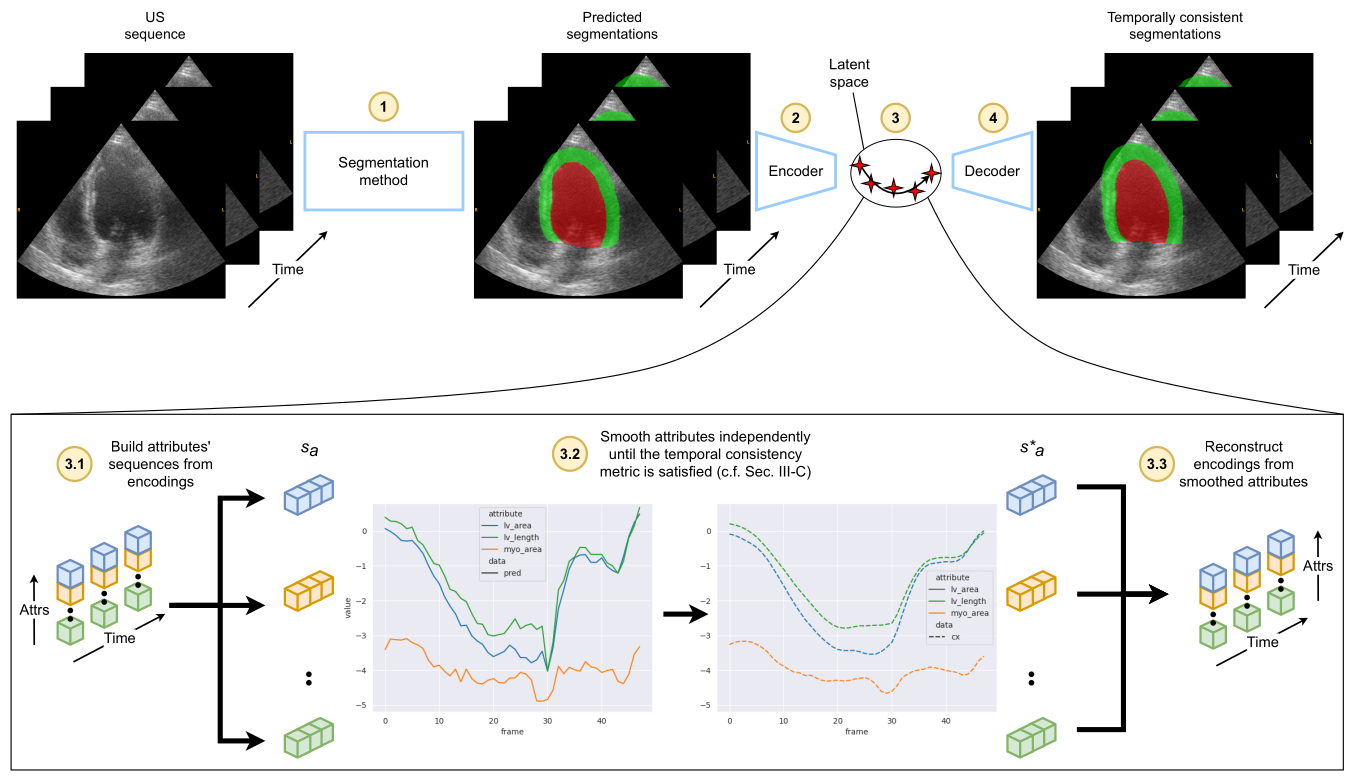
The figure below shows a schematic representation of our temporal regularization method. Starting with the raw echocardiography sequence, a SOTA segmentation method predicts a segmentation mask (1), which is then encoded frame-by-frame (2) by a pretrained autoencoder. There, the encodings of the sequence are split between dimensions (3.1) to produce sequences of attributes with respect to time (sa). These sequences are then processed invidually (3.2), and the results (s∗a) are merged back together as encodings for each frame (3.3). Finally, the modified encodings are decoded into now temporally consistent segmentations (4).
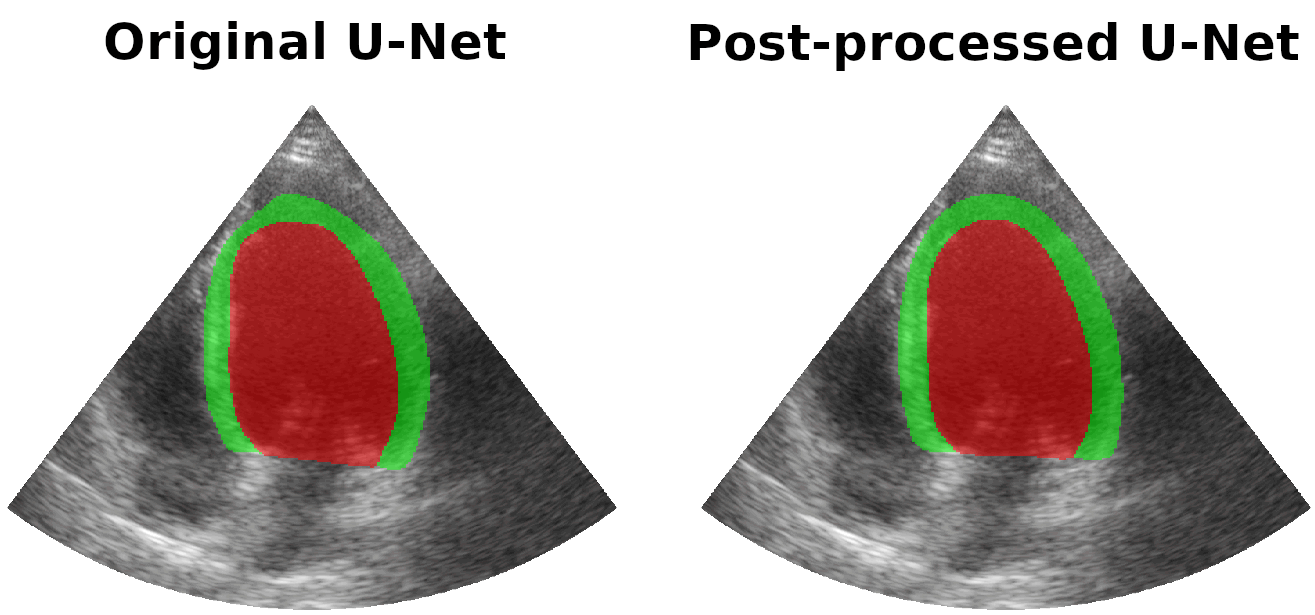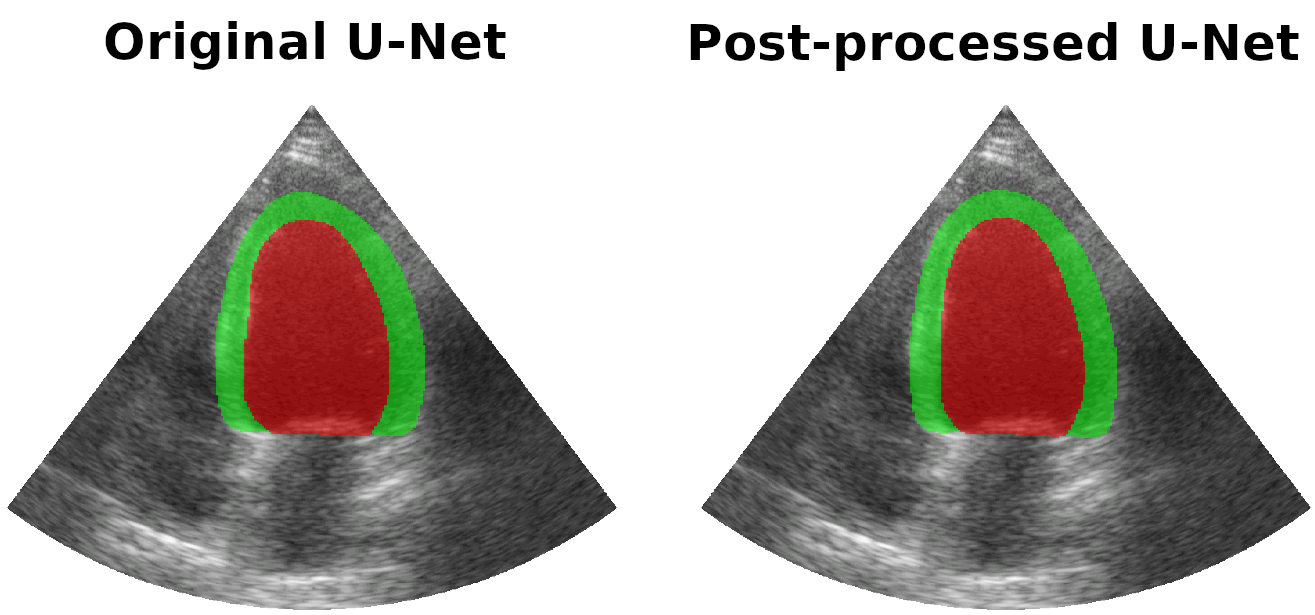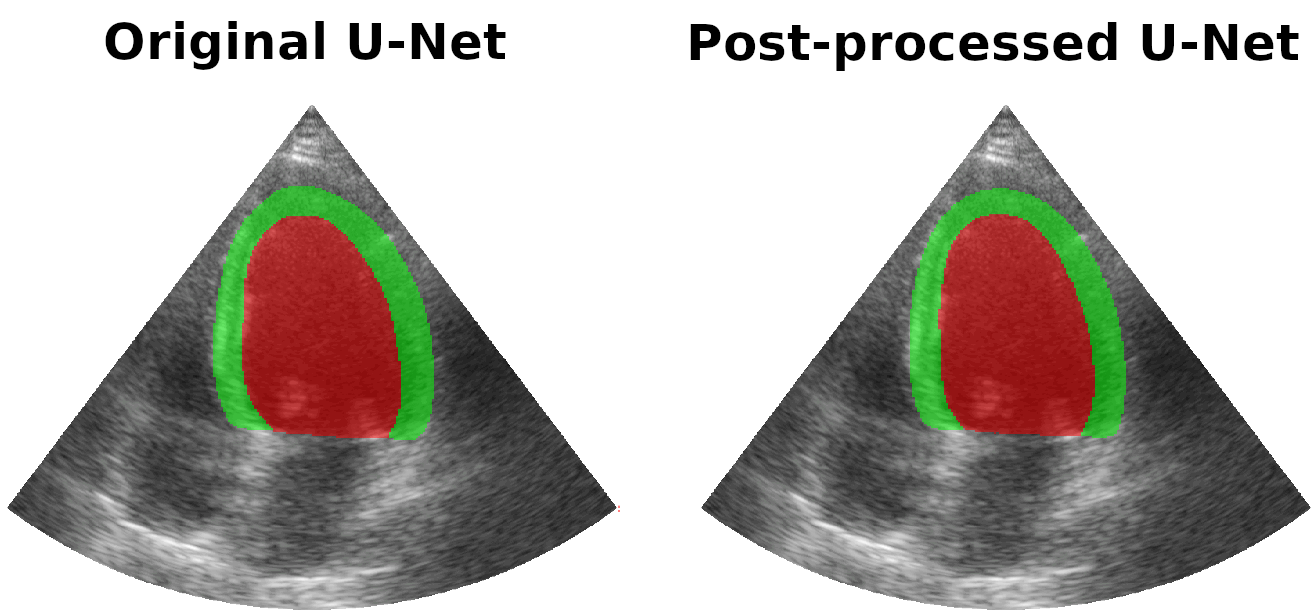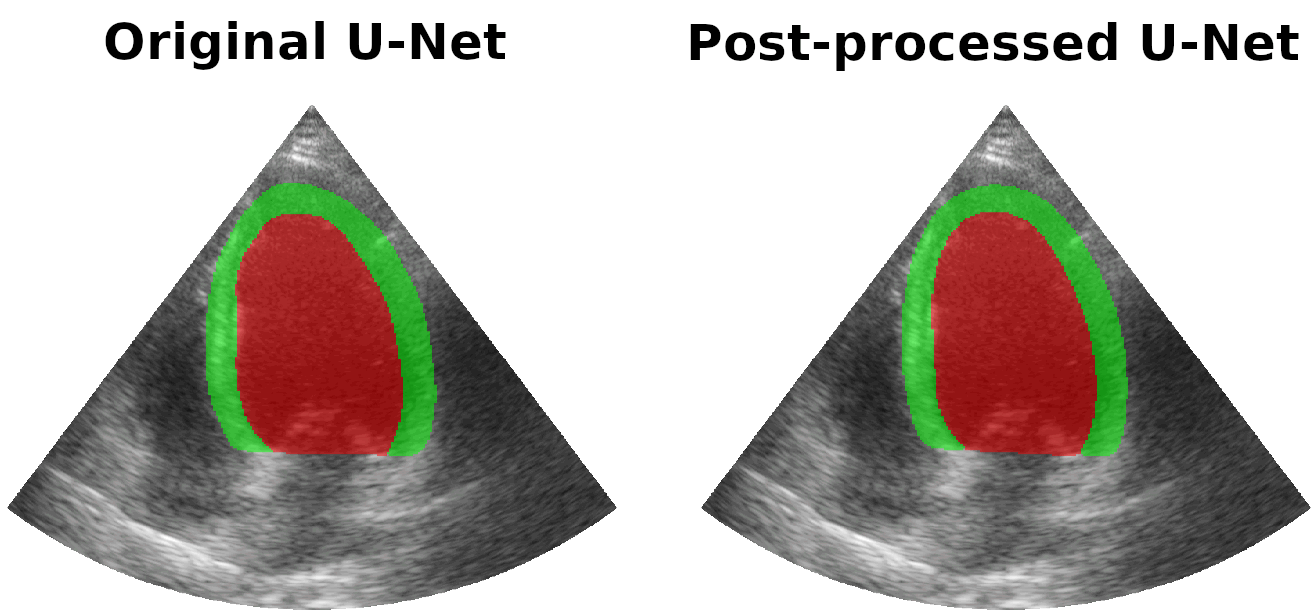
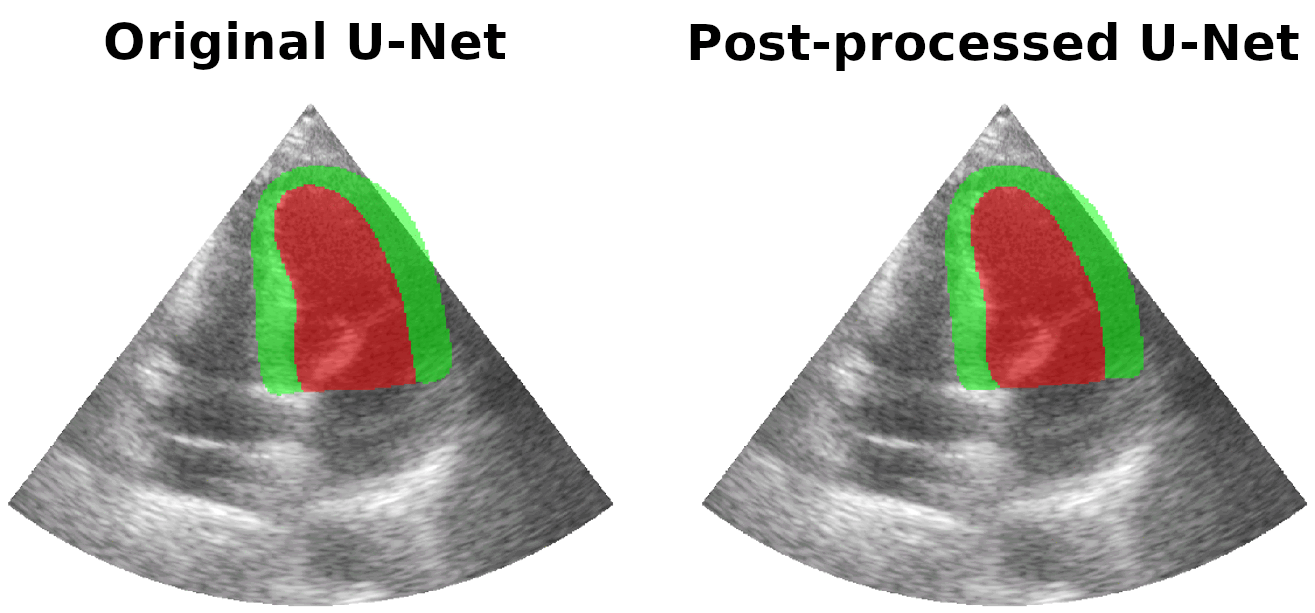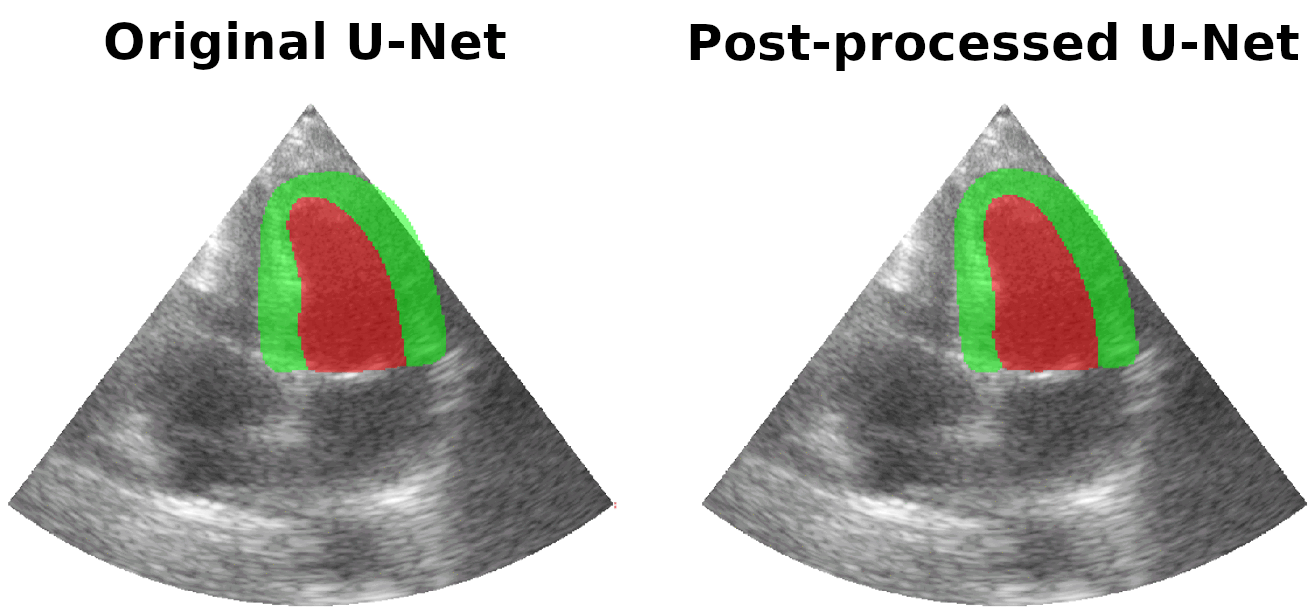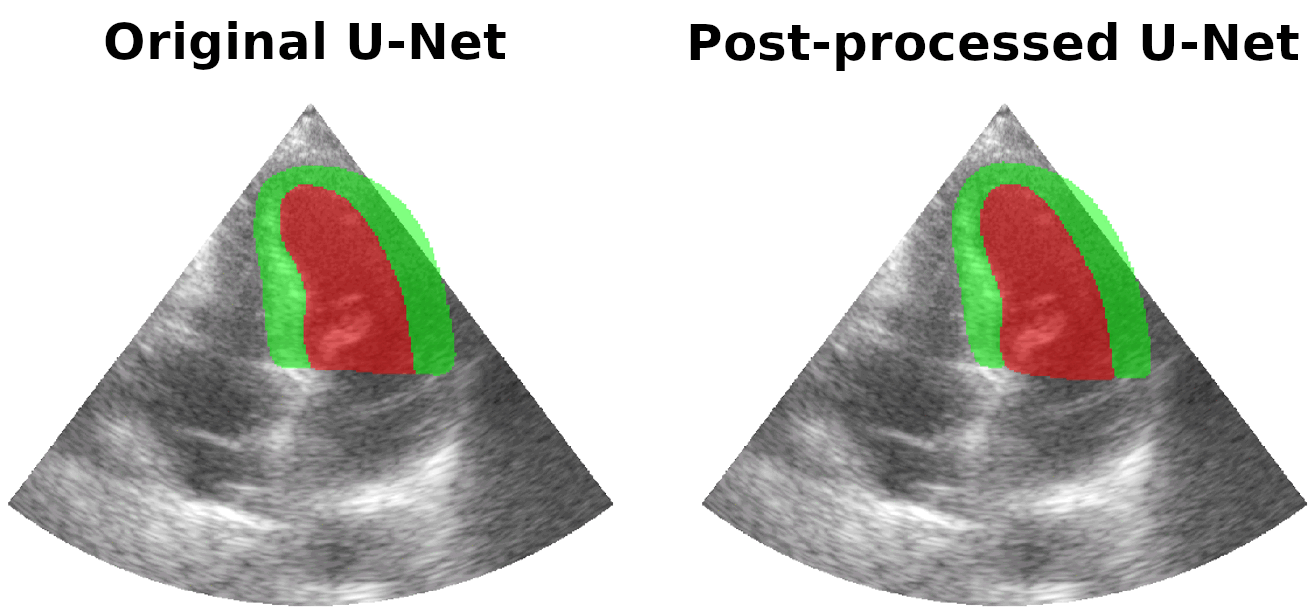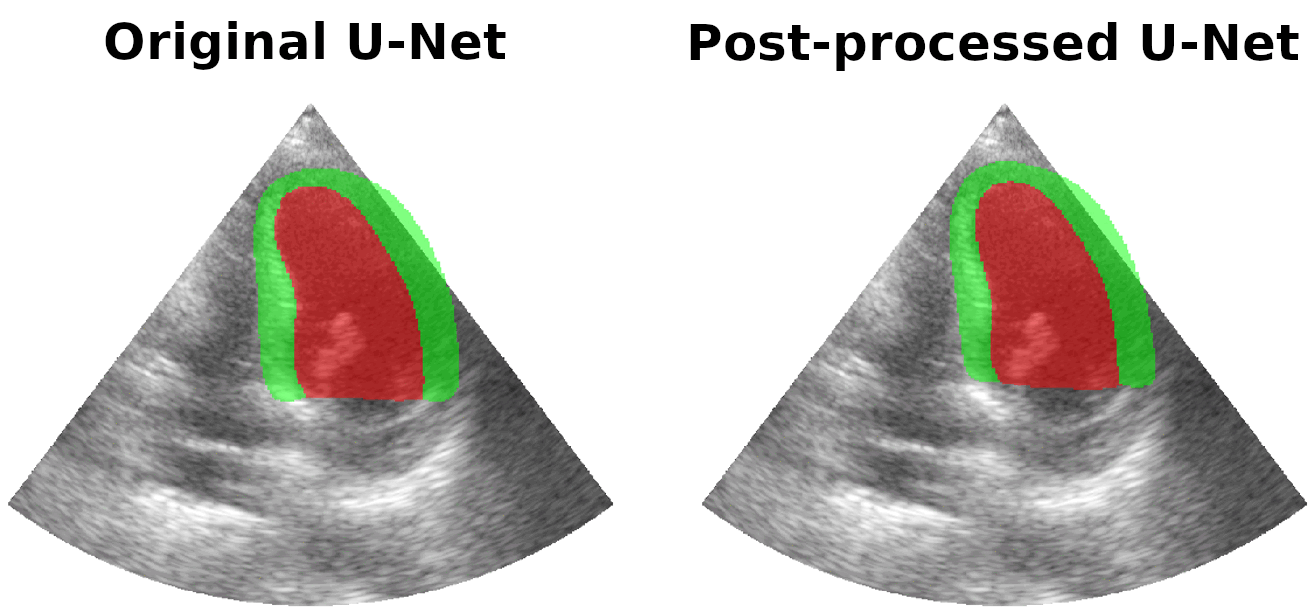
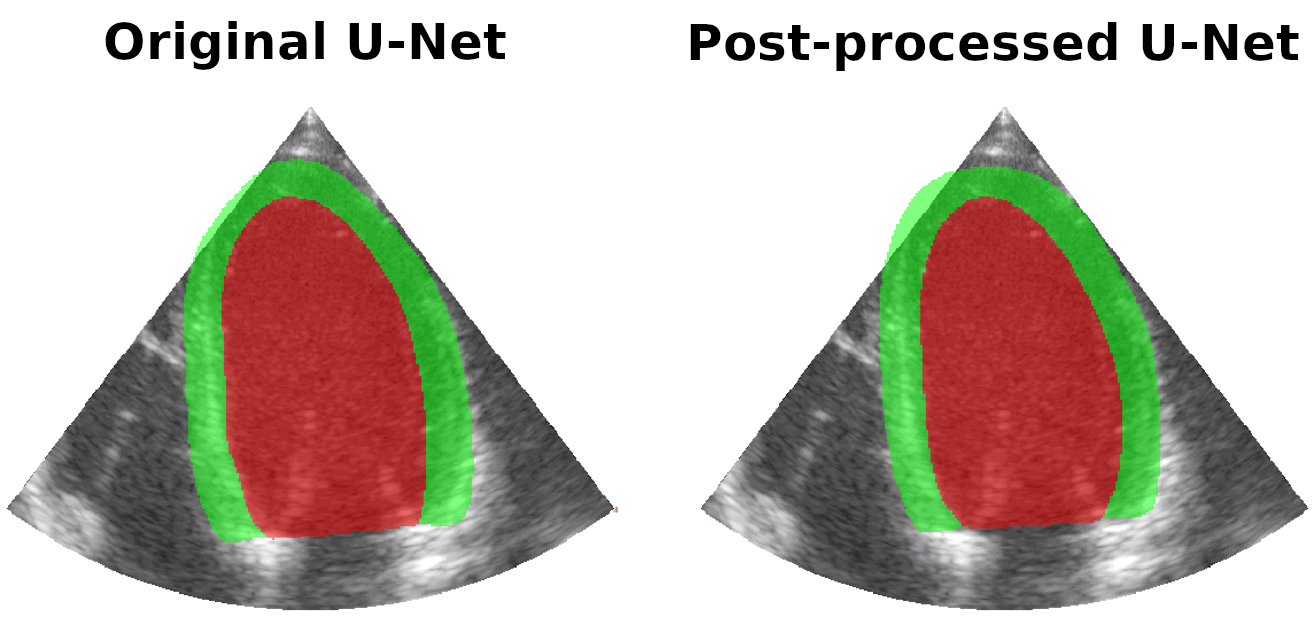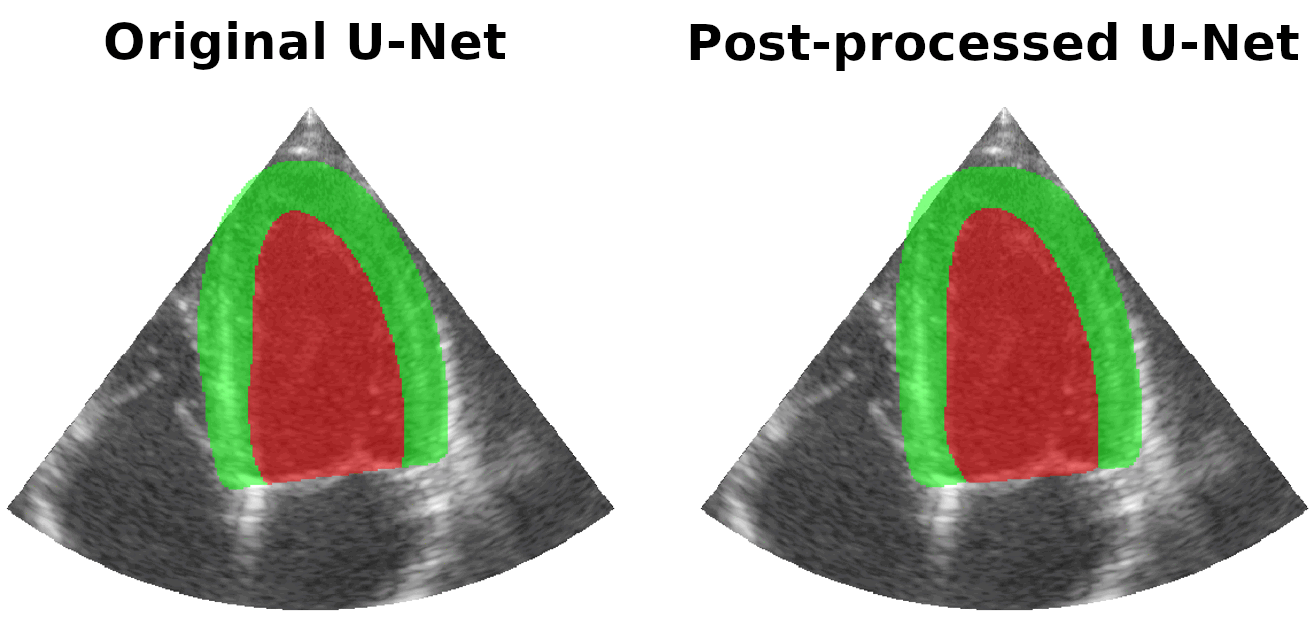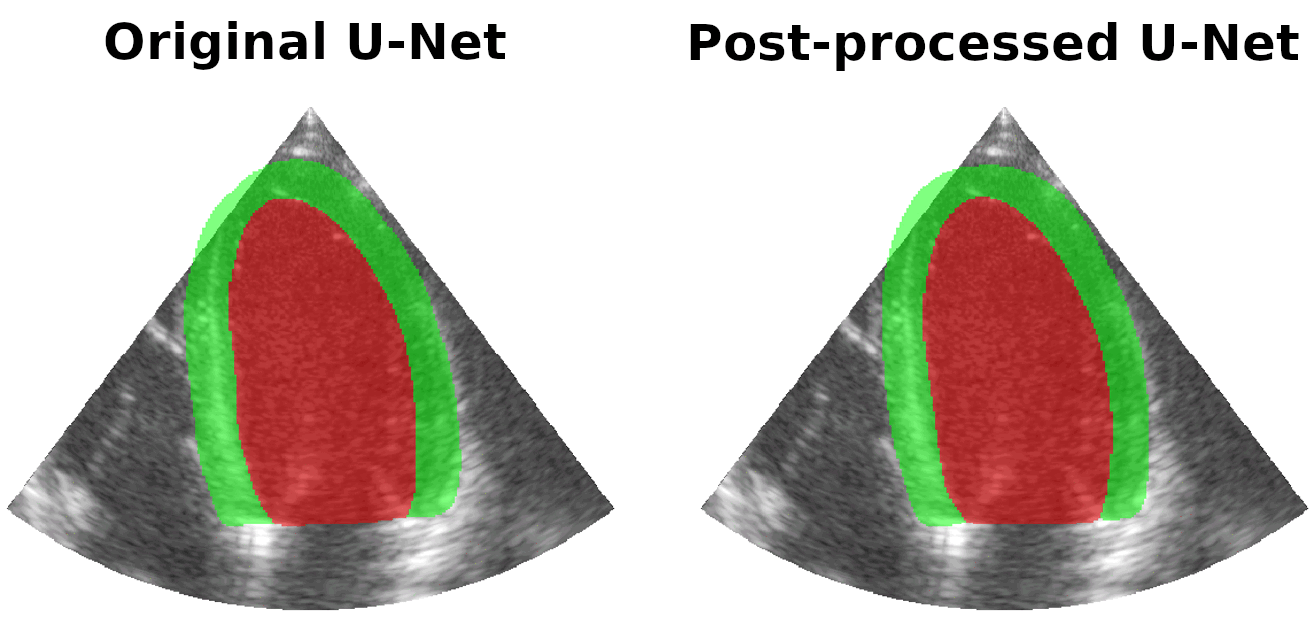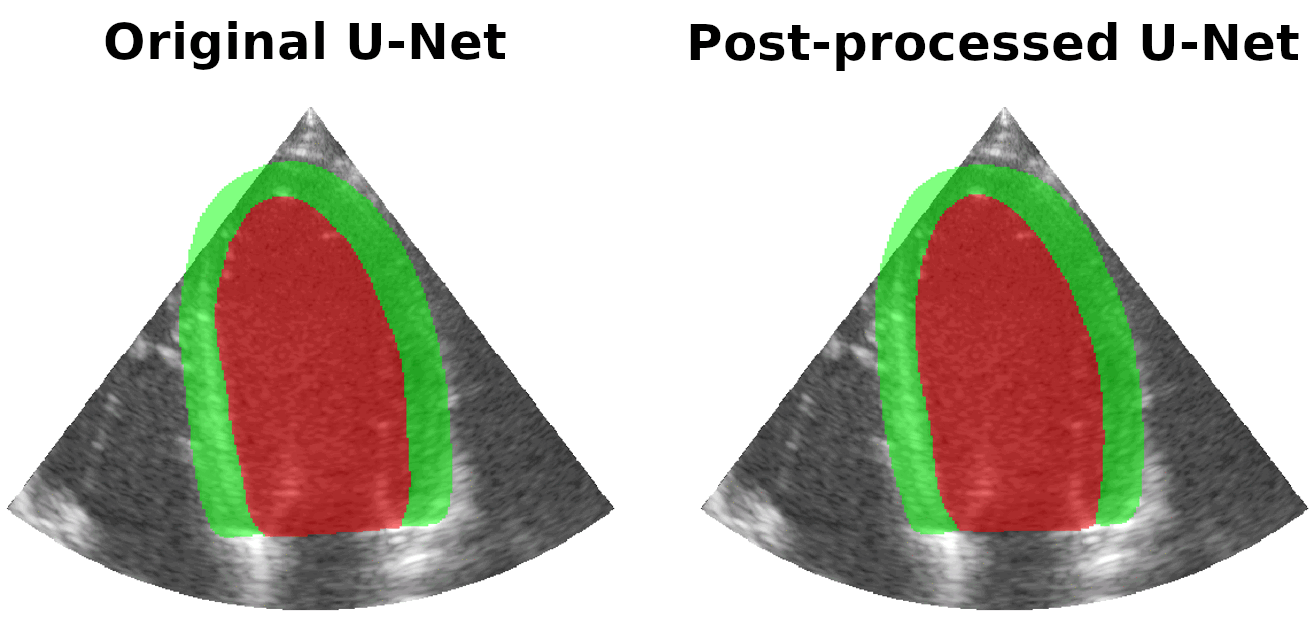
Results on real patients
For all animations, the superimposed masks correspond to those estimated by a standard U-Net method (left) which was then post-processed by our method (right). Don't hesitate to download each animation by right clicking on it for a better visualization.
R&D Team
| Nathan PAINCHAUD | PhD student, VITALab (Canada) and
CREATIS (France)
|
| Nicolas DUCHATEAU | Associate professor, CREATIS, France |
| Olivier BERNARD | Full professor, CREATIS, France
|
| Pierre-Marc JODOIN | Full professor, VITALab, Canada
|










